Genus¶
Overview¶
Genus statistics provide a measure of a region’s topology. At a given value in the data, the genus value is the number of discrete region above the value minus the number of regions below it. When this process is repeated over a range of values, a Genus curve can be constructed. The technique has previously been used to study CMB deviations from a Gaussian distribution.
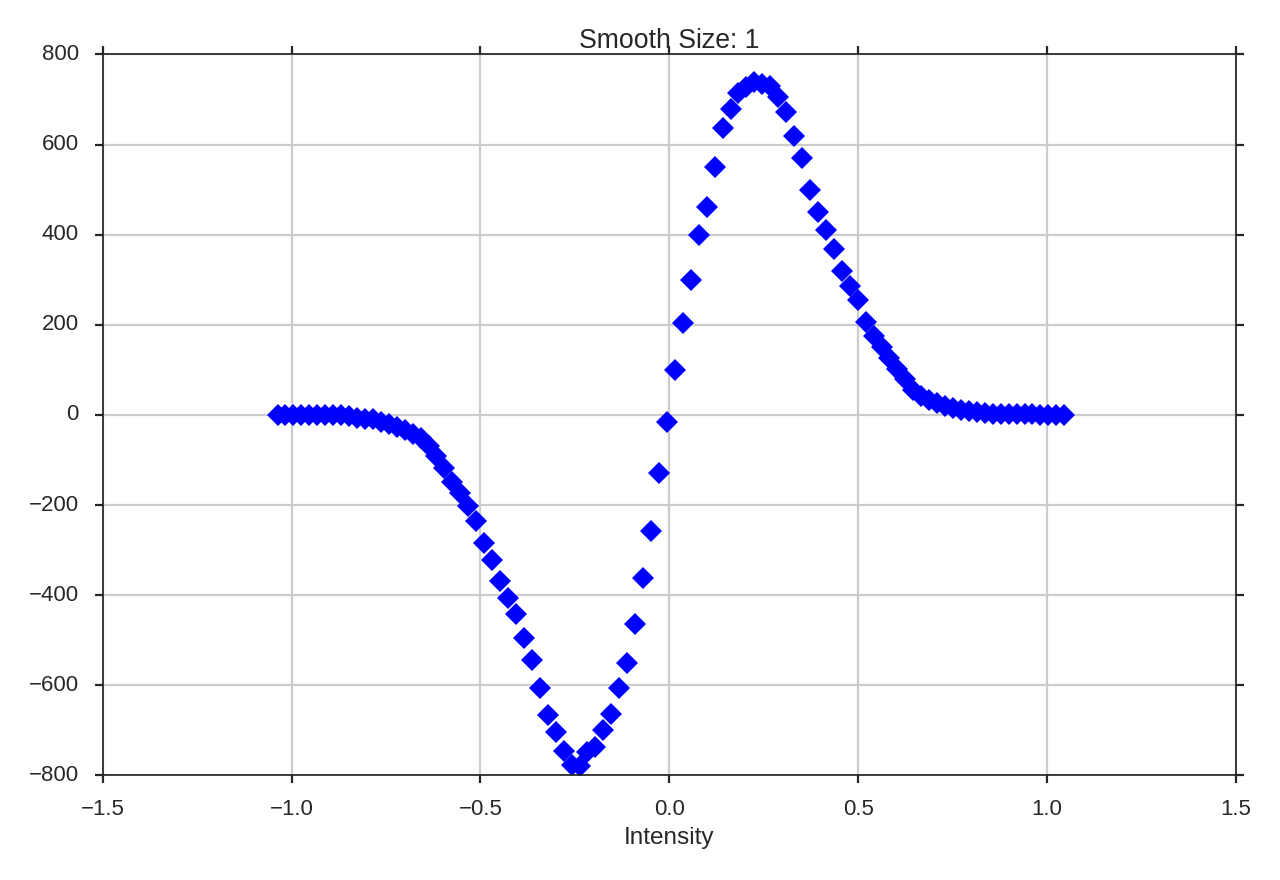
If a region has a negative Genus statistics, it is dominate by holes in the emission (“swiss cheese” morphology). A positive Genus value implies as “meatball” morphology, where the emission is localized into clumps. The Genus curve of a Gaussian field is shown below. Note that at the mean value (0.0), the Genus value is zero: at the mean intensity, there is no preference to either morphological type.
Kowal et al. 2007 constructed Genus curves for a set of simulations to investigate the effect of changing the Mach number and the Alfvenic Mach number. The isocontours were taken for a range of density values in the full position-position-position space.
Chepurnov et al. 2008 then used this technique on an HI integrated intensity image of the Small Magellanic Cloud (SMC). The range of values used to create the curve are the HI intensities in the image. They investigated the change in the morphology over several regions and at different smoothing scales.
Using¶
The data in this tutorial are available here.
We need to import the Genus code, along with a few other common packages:
>>> from turbustat.statistics import Genus
>>> from astropy.io import fits
>>> import astropy.units as u
>>> import numpy as np
And we load in the data:
>>> moment0 = fits.open("Design4_21_0_0_flatrho_0021_13co.moment0.fits")[0]
The FITS HDU is passed to initialize Genus:
>>> genus = Genus(moment0, lowdens_percent=15, highdens_percent=85, numpts=100, smoothing_radii=np.linspace(1, moment0.shape[0] / 10., 5))
lowdens_percent and highdens_percent set the upper and lower percentiles in the data to measure the Genus value at. When using observational data, lowdens_percent must be set above the noise level (construct a CDF if you are unsure; see turbustat.statistics.PDF for CDF construction). The numpts parameter sets how many values to compute the Genus value between the given percentiles. Finally, smoothing_radii allows for the data to be smoothed, minimizing the influence of noise on the Genus curve at the expense of resolution. The values given are used as the radii of a Gaussian smoothing kernel. The values given above (np.linspace(1, moment0.shape[0] / 10., 5)) are used by default when no values are given.
Computing the curves is accomplished using run:
>>> genus.run(verbose=True, min_npix=4)
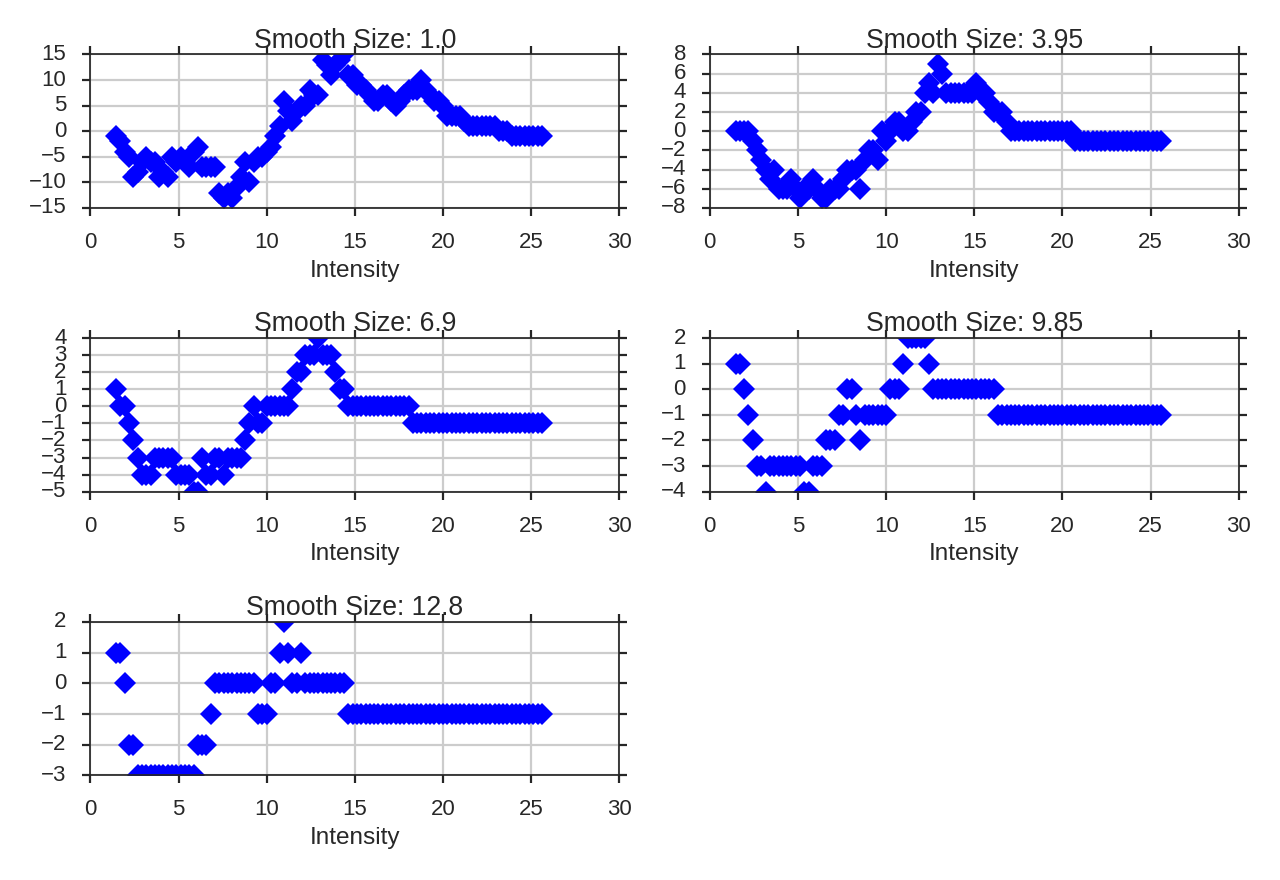
The basic sinusoid seen in the Genus curve of the Gaussian field is still evident. As we smooth the data on larger scales, the topological information is lost, and the curve becomes degraded. To avoid spurious noise features, the minimum number of pixels a region must have to be counted can be set by min_npix. This is simulated data, so a small value has been chosen.
Often the smallest size that can be “trusted” in a radio image is the beam area. In this example, a FITS HDU was passed, including an associated header. If the beam information is contained in the header, the size threshold can be set to the beam area using:
>>> genus = Genus(moment0, lowdens_percent=15, highdens_percent=85, smoothing_radii=[1])
>>> moment0.header["BMAJ"] = 2e-5 # deg.
>>> genus.run(verbose=True, use_beam=True)
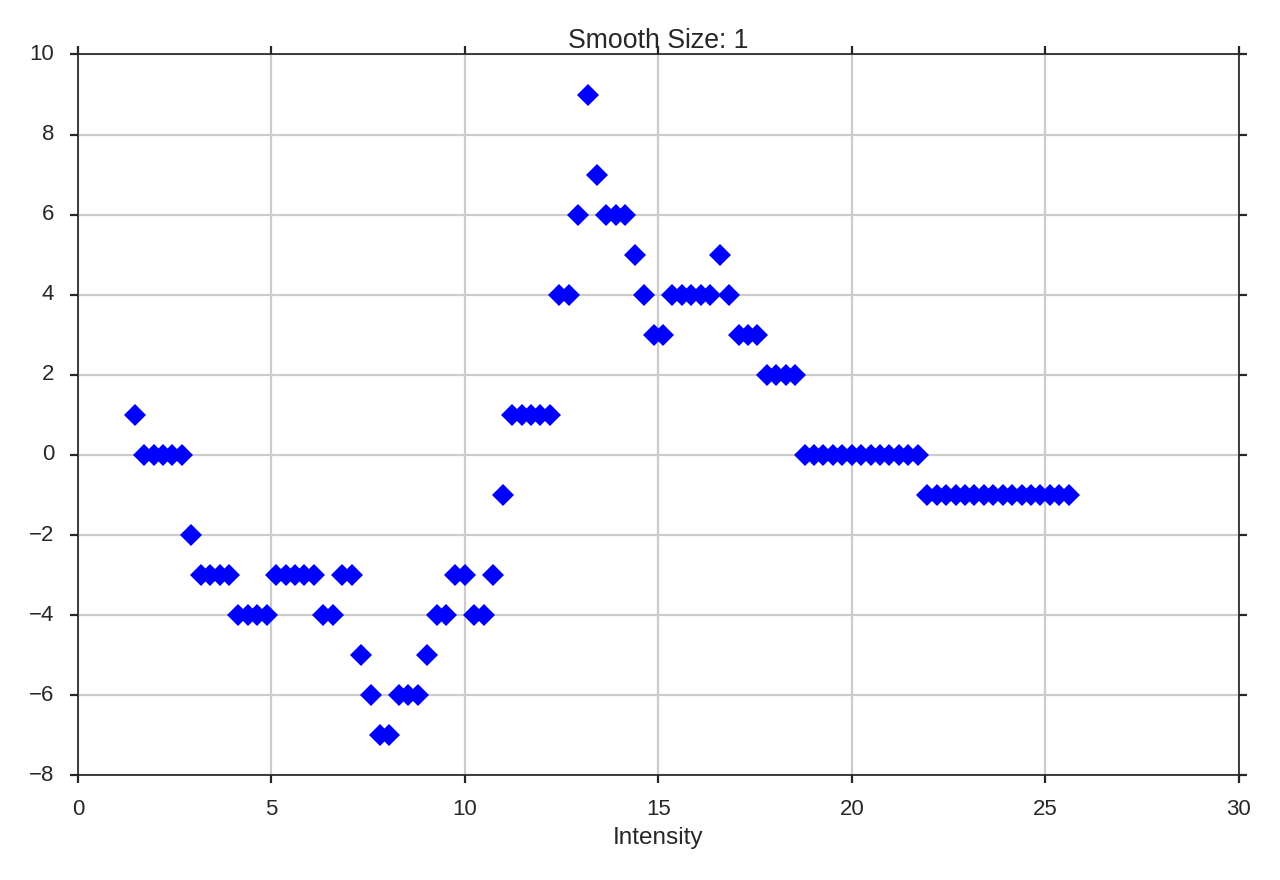
The curve has far less detail then before when requiring large connected regions. Note that the FITS keywords “BMIN” and “BPA” are also read and used, when available. More options for reading beam information are available when the optional package radio_beam is installed. If the beam information is not contained in the header, a custom area can be passed using beam_area,
>>> genus.run(verbose=True, use_beam=True, beam_area=2e-5**2 * np.pi * u.deg**2)
This returns the same result shown above.